

Our results on 12 signaling pathways from the NetPath database indicate that identifying the strongest path is helpful for pathway reconstruction. This allows StrongestPath to be used for any organism, PPI, gene regulatory networks, and signal transduction networks.

In addition, users can provide their own networks and nomenclature datasets. StrongestPath comes with two types of built-in databases: (I) some PPI and signaling networks of human and mouse, containing interactions recorded in public databases, (II) protein nomenclature database, containing 11 different symbols and accession IDs of genes and proteins in different databases. For example, when a list of genes is identified in a study of a phenomenon, researchers seek to answer whether there is a regulatory pathway between the transcription factors associated with the phenomenon and the identified genes regarding the experimentally validated data reported in the public databases. The third challenge is identifying any activating or inhibitory regulatory path between two distinct groups of proteins.

This feature can be used to identify unknown elements of a protein complex, biological process or core regulatory circuitry. To address this challenge, StrongestPath looks at the whole PPI or signaling network and identifies proteins with maximum total confidence of interactions with the given set of proteins. The second challenge addressed by StrongestPath is growing the sub-network of the input proteins, either by extracting their pairwise interactions from a list of PPI or signaling databases, or adding further proteins that are more likely to create protein complexes or dense interactions with the input proteins. In many experimental studies perturbation of a protein A is observed to influence a protein B, but the cascade of interactions between A and B is unidentified. The first challenge is identifying a cascade of interactions, as a regulatory or signaling pathway, in a large PPI or signaling network. We developed StrongestPath, a Cytoscape 3.0 App, to address three key challenges during analysis of PPI or signaling networks. PesCa, PathExplorer ( ) and PathLinker, are examples of such apps that can compute paths in biological networks. Other Plugins and Apps can be integrated into this flexible platform for complex network analysis and visualisations. To make this process easier, Cytoscape was developed to help with molecular and network profiling analysis and also visualizing molecular interaction networks.
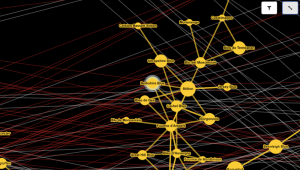
So, the identification of the cascades of interactions from the receptors to the transcriptional regulatory factors is a major challenge in systems biology. higher confidences for the experimentally validated interactions, and lower values for the computationally predicted ones. Some of the databases assign different confidence levels to the interactions, e.g. There have been numerous protein–protein interaction (PPI) or signaling pathways databases developed based on the experimental approaches or computational predictions. The teamwork of proteins, in terms of temporary or permanent interactions, is critical for any biological process. ConclusionĮasy access to multiple public large databases, generating output in a short time, addressing some key challenges in one platform, and providing a user-friendly graphical interface make StrongestPath an extremely useful application. Our results on 12 signaling pathways from the NetPath database demonstrate that the application can be used for indicating proteins which may play significant roles in a pathway by finding the strongest path(s) in the PPI or signaling network. This application can be used on the built-in human and mouse PPI or signaling databases, or any user-provided database for some organism. The application can also identify any activating or inhibitory regulatory paths between two distinct sets of transcription factors and target genes. Given a set of proteins, StrongestPath can extract a set of possible interactions between the input proteins, and expand the network by adding new proteins that have the most interactions with highest total confidence to the current network of proteins. When there are different levels of confidence over the interactions, the application is able to process them and identify the cascade of interactions with the highest total confidence score. StrongestPath is a Cytoscape 3 application that enables the analysis of interactions between two proteins or groups of proteins in a collection of protein–protein interaction (PPI) network or signaling network databases.


 0 kommentar(er)
0 kommentar(er)
